Accueil
actualités
mercredi 12 mars 2025
actualités
mardi 10 décembre 2024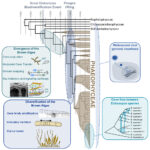
actualités
mercredi 13 novembre 2024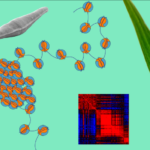
actualités
lundi 11 novembre 2024
actualités
lundi 11 novembre 2024
actualités
lundi 11 novembre 2024
actualités
dimanche 31 mars 2024
L'unité en quelques chiffres clés :
|
Créé en 2022 |
75 membres |
5 équipes de recherche |
308 publications (2015-2025) |
45 soutenances de thèse (2015-2025) |
Les équipes :
Publications récentes :
Bull, Emma C.; Singh, Archana; Harden, Amy M.; Soanes, Kirsty; Habash, Hala; Toracchio, Lisa; Carrabotta, Marianna; Schreck, Christina; Shah, Karan M.; Riestra, Paulina Velasco; Chantoiseau, Margaux; Costa, Maria Eugénia Marques Da; Moquin-Beaudry, Gaël; Pantziarka, Pan; Essiet, Edidiong Akanimo; Gerrand, Craig; Gartland, Alison; Bojmar, Linda; Fahlgren, Anna; Marchais, Antonin; Papakonstantinou, Evgenia; Tomazou, Eleni M.; Surdez, Didier; Heymann, Dominique; Cidre-Aranaz, Florencia; Fromigue, Olivia; Sexton, Darren W.; Herold, Nikolas; Grünewald, Thomas G. P.; Scotlandi, Katia; Nathrath, Michaela; Green, Darrell
Targeting metastasis in paediatric bone sarcomas Article de journal
Dans: Mol Cancer, vol. 24, no. 1, 2025, ISSN: 1476-4598.
@article{Bull2025,
title = {Targeting metastasis in paediatric bone sarcomas},
author = {Emma C. Bull and Archana Singh and Amy M. Harden and Kirsty Soanes and Hala Habash and Lisa Toracchio and Marianna Carrabotta and Christina Schreck and Karan M. Shah and Paulina Velasco Riestra and Margaux Chantoiseau and Maria Eugénia Marques Da Costa and Gaël Moquin-Beaudry and Pan Pantziarka and Edidiong Akanimo Essiet and Craig Gerrand and Alison Gartland and Linda Bojmar and Anna Fahlgren and Antonin Marchais and Evgenia Papakonstantinou and Eleni M. Tomazou and Didier Surdez and Dominique Heymann and Florencia Cidre-Aranaz and Olivia Fromigue and Darren W. Sexton and Nikolas Herold and Thomas G. P. Grünewald and Katia Scotlandi and Michaela Nathrath and Darrell Green},
doi = {10.1186/s12943-025-02365-z},
issn = {1476-4598},
year = {2025},
date = {2025-12-00},
urldate = {2025-12-00},
journal = {Mol Cancer},
volume = {24},
number = {1},
publisher = {Springer Science and Business Media LLC},
abstract = {<jats:title>Abstract</jats:title>
<jats:p>Paediatric bone sarcomas (e.g. Ewing sarcoma, osteosarcoma) comprise significant biological and clinical heterogeneity. This extreme heterogeneity affects response to systemic therapy, facilitates inherent and acquired drug resistance and possibly underpins the origins of metastatic disease, a key component implicit in cancer related death. Across all cancers, metastatic models have offered competing accounts on when dissemination occurs, either early or late during tumorigenesis, whether metastases at different foci arise independently and directly from the primary tumour or give rise to each other, i.e. metastases-to-metastases dissemination, and whether cell exchange occurs between synchronously growing lesions. Although it is probable that all the above mechanisms can lead to metastatic disease, clinical observations indicate that distinct modes of metastasis might predominate in different cancers. Around 70% of patients with bone sarcoma experience metastasis during their disease course but the fundamental molecular and cell mechanisms underlying spread are equivocal. Newer therapies such as tyrosine kinase inhibitors have shown promise in reducing metastatic relapse in trials, nonetheless, not all patients respond and 5-year overall survival remains at ~ 50%. Better understanding of potential bone sarcoma biological subgroups, the role of the tumour immune microenvironment, factors that promote metastasis and clinical biomarkers of prognosis and drug response are required to make progress. In this review, we provide a comprehensive overview of the approaches to manage paediatric patients with metastatic Ewing sarcoma and osteosarcoma. We describe the molecular basis of the tumour immune microenvironment, cell plasticity, circulating tumour cells and the development of the pre-metastatic niche, all required for successful distant colonisation. Finally, we discuss ongoing and upcoming patient clinical trials, biomarkers and gene regulatory networks amenable to the development of anti-metastasis medicines.</jats:p>},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
<jats:p>Paediatric bone sarcomas (e.g. Ewing sarcoma, osteosarcoma) comprise significant biological and clinical heterogeneity. This extreme heterogeneity affects response to systemic therapy, facilitates inherent and acquired drug resistance and possibly underpins the origins of metastatic disease, a key component implicit in cancer related death. Across all cancers, metastatic models have offered competing accounts on when dissemination occurs, either early or late during tumorigenesis, whether metastases at different foci arise independently and directly from the primary tumour or give rise to each other, i.e. metastases-to-metastases dissemination, and whether cell exchange occurs between synchronously growing lesions. Although it is probable that all the above mechanisms can lead to metastatic disease, clinical observations indicate that distinct modes of metastasis might predominate in different cancers. Around 70% of patients with bone sarcoma experience metastasis during their disease course but the fundamental molecular and cell mechanisms underlying spread are equivocal. Newer therapies such as tyrosine kinase inhibitors have shown promise in reducing metastatic relapse in trials, nonetheless, not all patients respond and 5-year overall survival remains at ~ 50%. Better understanding of potential bone sarcoma biological subgroups, the role of the tumour immune microenvironment, factors that promote metastasis and clinical biomarkers of prognosis and drug response are required to make progress. In this review, we provide a comprehensive overview of the approaches to manage paediatric patients with metastatic Ewing sarcoma and osteosarcoma. We describe the molecular basis of the tumour immune microenvironment, cell plasticity, circulating tumour cells and the development of the pre-metastatic niche, all required for successful distant colonisation. Finally, we discuss ongoing and upcoming patient clinical trials, biomarkers and gene regulatory networks amenable to the development of anti-metastasis medicines.</jats:p>
Rousselot, Ellyn; Nehr, Zofia; Aury, Jean-Marc; Denoeud, France; Cock, J Mark; Tirichine, Leïla; Duc, Céline
Identification of novel H2A histone variants across diverse clades of algae Article de journal
Dans: Genome Biology, vol. 26, p. 299, 2025.
@article{Rousselot2025,
title = {Identification of novel H2A histone variants across diverse clades of algae},
author = {Ellyn Rousselot and Zofia Nehr and Jean-Marc Aury and France Denoeud and J Mark Cock and Leïla Tirichine and Céline Duc},
editor = {Springer Nature},
year = {2025},
date = {2025-09-23},
urldate = {2025-06-18},
journal = {Genome Biology},
volume = {26},
pages = {299},
abstract = {Background
Histones are among the most conserved proteins in eukaryotes. They not only ensure DNA compaction in the nucleus but also participate in epigenetic regulation of gene expression. These key epigenetic players are divided into replication-coupled histones, expressed during the S-phase, and replication-independent variants, expressed throughout the cell cycle. Compared with other core histones, H2A proteins exhibit a high level of variability but the characterization of algal H2A variants remains very limited.
Results
In this study, we exploit genome and transcriptome data from 22 species to identify H2A variants in brown seaweeds. Combined analyses of phylogenetic data, synteny and protein motifs enable us to reveal the presence of new H2A variants as well as their evolutionary history. We identify three new H2A variants: H2A.N, H2A.O and H2A.E. In brown seaweeds, the H2A.E and H2A.O variants arose from the same monophyletic clade while the H2A.N variant emerged independently. Moreover, the H2A.E variant seems to have a shared ancestry with RC H2A while the H2A.O variant has an H2A.X-characteristic signature without being orthologous to this variant. Based on mass spectrometry, we identify distinct epigenetic marks on these H2A variants. Finally, the H2A.Z, H2A.N and H2A.O from brown seaweeds are ubiquitously expressed while expression of H2A.E has tissue-specific patterns, especially in reproductive tissues.
Conclusions
We thus hypothesize that H2A.O and H2A.X might have convergent functions while H2A.E might fulfil some functions of replication-coupled H2As and/or compensate for the absence of repressive histone marks along with H2A.N.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Histones are among the most conserved proteins in eukaryotes. They not only ensure DNA compaction in the nucleus but also participate in epigenetic regulation of gene expression. These key epigenetic players are divided into replication-coupled histones, expressed during the S-phase, and replication-independent variants, expressed throughout the cell cycle. Compared with other core histones, H2A proteins exhibit a high level of variability but the characterization of algal H2A variants remains very limited.
Results
In this study, we exploit genome and transcriptome data from 22 species to identify H2A variants in brown seaweeds. Combined analyses of phylogenetic data, synteny and protein motifs enable us to reveal the presence of new H2A variants as well as their evolutionary history. We identify three new H2A variants: H2A.N, H2A.O and H2A.E. In brown seaweeds, the H2A.E and H2A.O variants arose from the same monophyletic clade while the H2A.N variant emerged independently. Moreover, the H2A.E variant seems to have a shared ancestry with RC H2A while the H2A.O variant has an H2A.X-characteristic signature without being orthologous to this variant. Based on mass spectrometry, we identify distinct epigenetic marks on these H2A variants. Finally, the H2A.Z, H2A.N and H2A.O from brown seaweeds are ubiquitously expressed while expression of H2A.E has tissue-specific patterns, especially in reproductive tissues.
Conclusions
We thus hypothesize that H2A.O and H2A.X might have convergent functions while H2A.E might fulfil some functions of replication-coupled H2As and/or compensate for the absence of repressive histone marks along with H2A.N.
Landry, Camille; Klusa, Daria; Cochonneau, Denis; Supiot, Stéphane; Heymann, Dominique
Circulating tumour cells for the prediction of the response to radiation therapy in prostate cancer Article de journal
Dans: 2025, ISSN: 1748-880X.
@article{Landry2025,
title = {Circulating tumour cells for the prediction of the response to radiation therapy in prostate cancer},
author = {Camille Landry and Daria Klusa and Denis Cochonneau and Stéphane Supiot and Dominique Heymann},
doi = {10.1093/bjr/tqaf224},
issn = {1748-880X},
year = {2025},
date = {2025-09-13},
urldate = {2025-09-13},
publisher = {Oxford University Press (OUP)},
abstract = {<jats:title>Abstract</jats:title>
<jats:p>Circulating tumour cells (CTCs) have emerged as a promising biomarker for assessing prognosis and predicting therapeutic efficacy in various cancers, including metastatic prostate cancer. However, predicting patient response to treatment, including radiation therapy (RT), remains a significant clinical challenge. This review explores the value of CTCs as prognostic markers in RT for prostate cancer, discussing their detection methods, biological significance, clinical relevance, and future implications.</jats:p>},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
<jats:p>Circulating tumour cells (CTCs) have emerged as a promising biomarker for assessing prognosis and predicting therapeutic efficacy in various cancers, including metastatic prostate cancer. However, predicting patient response to treatment, including radiation therapy (RT), remains a significant clinical challenge. This review explores the value of CTCs as prognostic markers in RT for prostate cancer, discussing their detection methods, biological significance, clinical relevance, and future implications.</jats:p>
Chandola, Udita; Manirakiza, Eric; Maillard, Margaux; Lavier-Aydat, Louis-Josselin; Camuel, Alicia; Trottier, Camille; Tanaka, Astuko; Chaumier, Timothée; Giraud, Eric; Tirichine, Leila
A Bradyrhizobium isolate from a marine diatom induces nitrogen-fixing nodules in a terrestrial legume Article de journal
Dans: Nature Microbiology, 2025.
@article{Chandola2025,
title = {A Bradyrhizobium isolate from a marine diatom induces nitrogen-fixing nodules in a terrestrial legume},
author = {Udita Chandola and Eric Manirakiza and Margaux Maillard and Louis-Josselin Lavier-Aydat and Alicia Camuel and Camille Trottier and Astuko Tanaka and Timothée Chaumier and Eric Giraud and Leila Tirichine},
editor = {Nature},
doi = {10.1038/s41564-025-02105-5},
year = {2025},
date = {2025-09-05},
journal = {Nature Microbiology},
abstract = {Biological nitrogen fixation converts atmospheric nitrogen into ammonia, essential to the global nitrogen cycle. While cyanobacterial diazotrophs are well characterized, recent studies have revealed a broad distribution of non-cyanobacterial diazotrophs (NCDs) in marine environments, although their study is limited by poor cultivability. Here we report a previously uncharacterized Bradyrhizobium isolated from the marine diatom Phaeodactylum tricornutum. Phylogenomic analysis places the strain within photosynthetic Bradyrhizobium, suggesting evolutionary adaptations to marine and terrestrial niches. Average nucleotide identity supports its classification as a previously undescribed species. Remarkably, inoculation experiments showed that the isolate induced nitrogen-fixing nodules in the Aeschynomene indica legume, pointing to symbiotic capabilities across ecological boundaries. Pangenome analysis and metabolic predictions indicate that this isolate shares more features with terrestrial photosynthetic Bradyrhizobium than with marine NCDs. Overall, these findings suggest that symbiotic interactions could evolve across different ecological niches, and raise questions about the evolution of nitrogen fixation and microbe–host interactions.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Dhingra, Surbhi; Téletchéa, Stéphane; Sowdhamini, Ramanathan; Sanejouand, Yves-Henri; Brevern, Alexandre G.; Cadet, Frédéric; Offmann, Bernard
Using protein blocks to build custom fragment libraries from protein structures Article de journal À paraître
Dans: Biochimie, À paraître, ISSN: 0300-9084.
@article{DHINGRA2025,
title = {Using protein blocks to build custom fragment libraries from protein structures},
author = {Surbhi Dhingra and Stéphane Téletchéa and Ramanathan Sowdhamini and Yves-Henri Sanejouand and Alexandre G. Brevern and Frédéric Cadet and Bernard Offmann},
url = {https://www.sciencedirect.com/science/article/pii/S0300908425001907},
doi = {https://doi.org/10.1016/j.biochi.2025.08.011},
issn = {0300-9084},
year = {2025},
date = {2025-08-13},
urldate = {2025-01-01},
journal = {Biochimie},
abstract = {The remarkable structural diversity of modern proteins reflects millions of years of evolution, during which sequence space has expanded while many structural features remain conserved. This conservation is evident not only among homologous proteins but also in the recurrence of supersecondary motifs across unrelated proteins, underscoring the abundance and robustness of these structural units. Here, we present a novel pipeline for generating customized protein fragment libraries using protein blocks (PBs)—a structural alphabet that encodes local backbone conformations. Our method efficiently extracts structurally similar fragments from a curated, non-redundant protein structure database by converting three-dimensional structures into one-dimensional PB sequences. By integrating predicted PB sequences with the PB-ALIGN and PB-kPRED tools, our approach identifies relevant fragments independently of sequence homology. Fragment quality is further assessed using a new scoring function that combines secondary structure similarity and PB alignment metrics. The resulting libraries contain fragments of at least seven PBs (11 amino acid residues), covering over 70% of the local backbone structure. Our results demonstrate that PBs enable efficient mining of high-quality structural fragments from diverse protein spaces, including proteins with disordered regions. The pipeline is accessible as an online tool (PB-Frag, http://pbpred-us2b.univ-nantes.fr/pbfrag).},
keywords = {},
pubstate = {forthcoming},
tppubtype = {article}
}